Home > Research > Research Overview
Just as Gregor Mendel followed the traits of pea plants to conclude that unseen “elements” (now known to be the DNA sequences that comprise individual genes) predictably determine those traits, so do we monitor specific traits in laboratory mice to identify the genes, and their defects, responsible for specific traits. Today, we are empowered by a far deeper understanding of genetics than held in Mendel’s time, and by immense advances in technology, enabling us to answer fundamental questions about the causes of human traits and diseases, from hair color to autoimmunity to Alzheimer’s disease.
The mouse as a model for humans
Genetic studies in a physiological setting require a model organism that can be experimentally manipulated in a controlled environment. The laboratory mouse serves as the premier model system for the study of mammalian biology. The anatomy and physiology of mice and humans are highly similar, with the same organs, organ systems, and organization within the body; most cell types and their functions are also shared. Ninety-nine percent of human genes have homologues in mice (ie, 99% of human and mouse genes have a shared ancestry), and 80% have orthologs (ie, 80% of human and mouse genes with shared ancestry have remained intact and unduplicated since their last common ancestor); in addition, 90% of the mouse genome exists in segments in which the gene order has been conserved with that in the human genome. Thus, discoveries made using mice usually have corresponding implications in humans.

Finding the genes responsible: the forward genetic approach
To fully understand any system, we must have a complete list of its parts and knowledge of how those parts work together. The approach employed in the Center for the Genetics of Host Defense aims to identify all necessary genes contributing to particular physiological processes. Termed “forward genetics,” this approach entails three essential steps: mutagenesis, the induction of heritable mutations in mice using the chemical mutagen N-ethyl-N-nitrosourea (ENU); phenotypic screening, examination and functional testing of the mutagenized mice and their cells for specific defects (for example, elevated blood glucose); and genetic mapping, identification of the mutated gene responsible for the defect (Figure 1).
Because no assumptions are made about which genes might be important for a biological process, the forward genetic approach is unbiased, and errors of interpretation are extremely rare. Moreover, the random nature of ENU mutagenesis means that no genes are overlooked. Eventually all genes will be tested, and those found necessary will encompass the parts list needed for full understanding of the process in question.
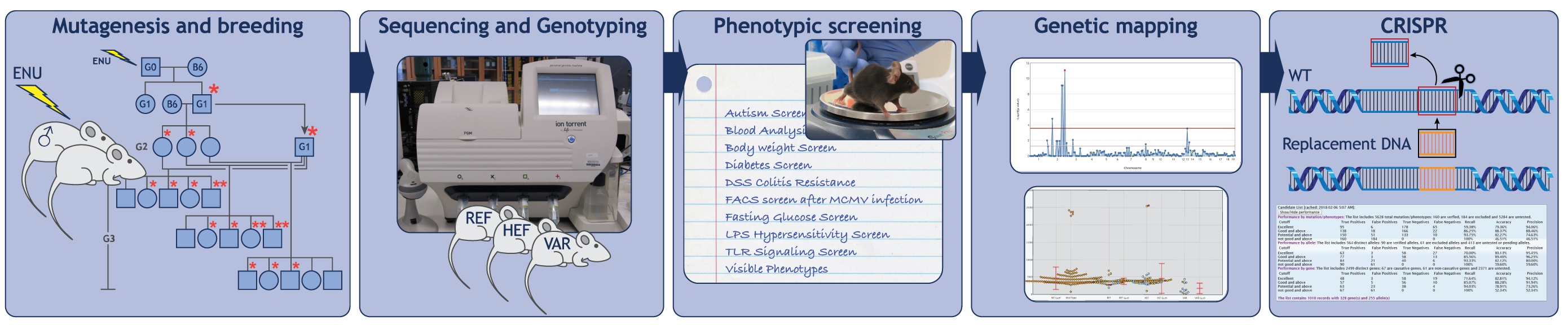
How genes work together: understanding mechanisms
Concurrent with obtaining a parts list, we work to determine the job of each part, and how the parts fit together to control characteristics of human biology, for example, blood glucose levels or susceptibility to a particular disease (Figure 2). We use a wide variety of methods in molecular biology, biochemistry, biophysics, structural biology, bioinformatics, and other disciplines to test hypotheses concerning the genes in question. As we continue to discover more genes necessary for a particular process, we gain insight into the mechanics of that process and can begin to understand the roles of individual genes. We publish our findings in top-tier peer-reviewed journals. We also directly post mutations, in both known and novel genes, to the Mutagenetix database.
Developing treatments for human disease
A full understanding of gene function, as described above, forms the basis for developing effective treatments for human disease. Against this backdrop of growing knowledge, we seek to identify novel drugs and drug targets that may provide new strategies to treat or cure disease. We pursue the development of these drugs and targets through collaborations with both academic and industry laboratories. Early work from the Beutler Laboratory (Peppel, Crawford, and Beutler. J Exp Med. 174, 1483-1489) led to the development of recombinant TNF inhibitors, a principle adopted in the development of Enbrel (Figure 3, a drug commonly used in the treatment of rheumatoid arthritis and other inflammatory diseases. The prototypic chimeric protein developed in the Beutler Laboratory consisted of the extracellular domain of the human p55 TNF receptor attached to the Fc portion and hinge region of a mouse IgG1 heavy chain. The protein forms a homodimer that binds and inactivates TNF.
Building upon our understanding of the Toll-like receptors, we have collaborated with the laboratories of Professor Dale Boger (The Scripps Research Institute) and Dr. Hong Zhang (UT Southwestern) to develop new adjuvants: drugs that enhance the immune response to give protection against infectious agents and cancerous cells (Innate Immune Drug Screening) (Video 1). This work entails screening synthetic compounds produced by chemists in the Boger Laboratory for the ability to stimulate the innate immune response in mouse and human macrophages, and in live mice. Those with stimulating activity are isolated and refined to produce highly active innate immune activators. Genetic and biochemical experiments are used to identify the targets of such compounds, and molecular modeling and X-ray crystallography are employed to understand interactions between the compound and its target. This knowledge permits further refinements to generate a potent and specific compound, which goes on to further testing in vivo for adjuvant and other effects.